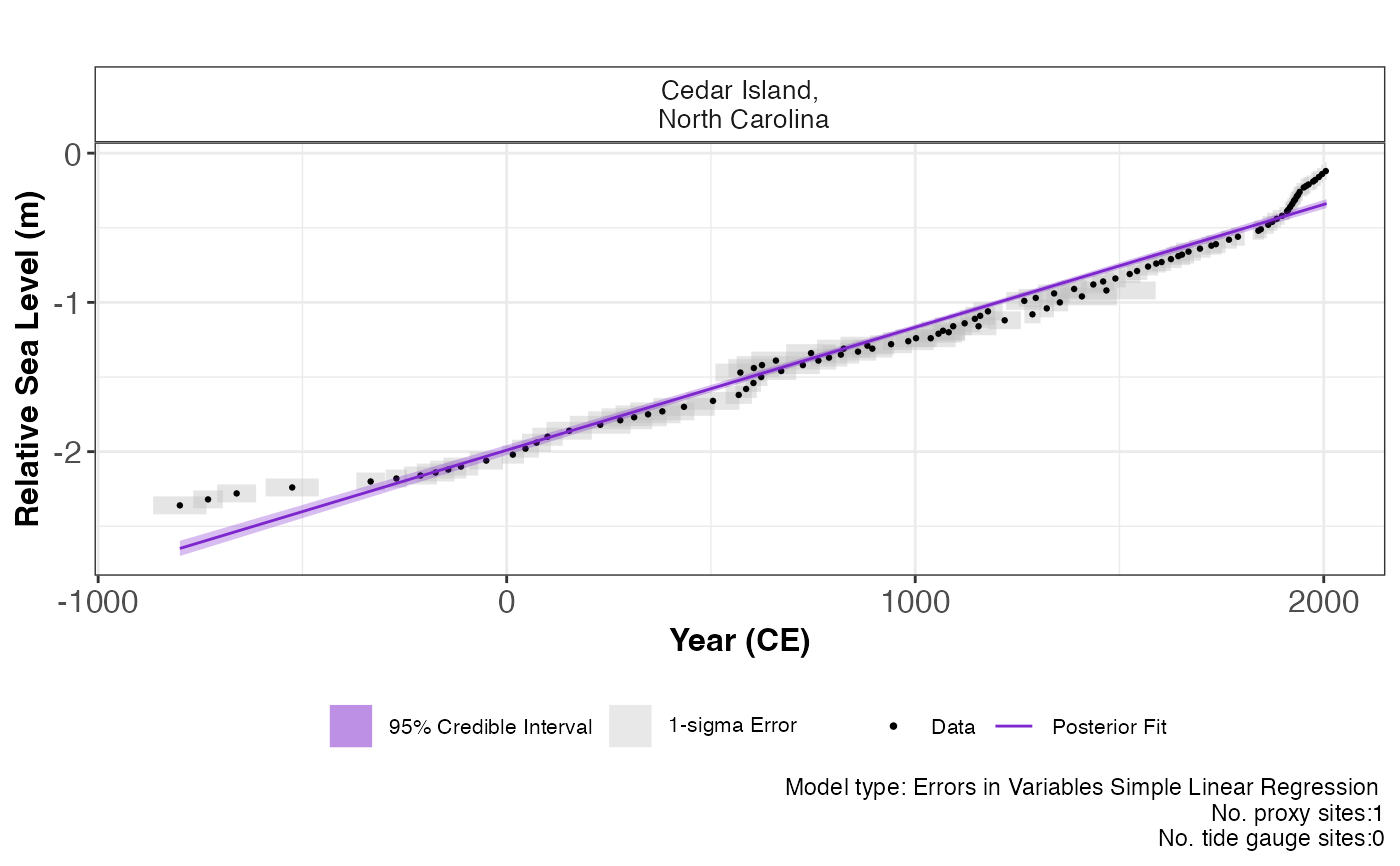
Plotting the results for each statistical model from the reslr_mcmc function.
Source: R/plot.reslr_output.R
plot.reslr_output.RdDepending on the model chosen in the reslr_mcmc function, the package produces a range of output plots.
Total posterior model fit plot with the raw data and measurement uncertainty are created for each statistical model.
The rate of change plots are created for the EIV IGP and the NI spline regression models.
For the NI GAM decomposition, each individual component of the model is plotted. Also, the regional and the non-linear local component, an associated rate plot is produced.
If tide gauges are used in the model, the user has the ability plot the output with or without this additional data source.
Arguments
- x
An object of class
reslr_outputandmodel_typecreated viareslr_mcmc- plot_proxy_records
Plotting the proxy records on their own and this is the default
- plot_tide_gauges
Plotting the tide gauge data as well as proxy data
- title
Plotting a title on the output plots
- plot_type
The user can select the type of output plot they require from the following: "rate_plot", "model_fit_plot", "regional_plot", "regional_rate_plot", "linear_local_plot", "non_linear_local_plot", "non_linear_local_rate_plot", "nigam_component_plot"
- plot_caption
Plotting an informed caption with the number of tide gauges and proxy sites.
- xlab
Labeling the x-axis
- ylab
Labeling the y-axis
- y_rate_lab
Labeling the y-axis for rate of change plots
- ...
Not used
Value
Plot of model fit and the rate of change depending on the statistical model in question.
Examples
# \donttest{
data <- NAACproxydata %>% dplyr::filter(Site == "Cedar Island")
x <- reslr_load(data = data)
jags_output <- reslr_mcmc(x, model_type = "eiv_slr_t")
#> Compiling model graph
#> Resolving undeclared variables
#> Allocating nodes
#> Graph information:
#> Observed stochastic nodes: 208
#> Unobserved stochastic nodes: 107
#> Total graph size: 1003
#>
#> Initializing model
#>
plot(x = jags_output)# }